CYBERSHUTTLE
Helping biophysicists spend 60% less time managing tools,
and more time advancing their research.

OVERVIEW
Cybershuttle helps biophysicists run molecular simulations without juggling multiple tools.
Biophysicists rely on disconnected tools, files, and supercomputing systems to conduct research. Managing this fragmented workflow often takes more effort than the scientific analysis itself.
Timeline
3 months (May 2023 - Aug 2023)
Team members
1 UX Designer (Self); 3 Developers; 1 Project Manager, 1 In-house scientist
My contribution
I led the UX research (Primary and secondary), ideation and UX design efforts (Low-fidelity and high-fidelity prototypes), and user testing to create an intuitive experience, addressing workflow challenges faced by biophysicists.
The Impact
60% ⬆
Faster research and experimentation.
42% ⬇
Reduced cognitive effort wrt scientific terminology.
3.5x
Faster project and team management.
THE PROBLEM
Biophysicists are frustrated when trying to execute and monitor experiments because their workflow is fragmented across different tools and devices.
MOLECULE PREP
BIOPHYSICISTS’ COMPUTER
BIOPHYSICISTS’ COMPUTER
SUPERCOMPUTER
EXPERIMENT SETUP
RUN SIMULATION
VISUALIZATION
ANALYSIS
THE PROCESS
I started with studying the biophysicists' ecosystem. I learnt about the different tools and devices they were using for each stage of their research process.
BIOPHYSICISTS’ COMPUTER
SUPERCOMPUTER
BIOPHYSICISTS’ COMPUTER
MOLECULE PREP
VMD
PyMOL
ChimeraX
EXPERIMENT SETUP
NAMD
GROMACS
CHARMM
RUN SIMULATION
SLURM
SSH
Azure
VISUALIZATION
VMD
PyMOL
ChimeraX
ANALYSIS
GROMACS
MDAnalysis
Excel/CSVs
SUPERCOMPUTER
Tools used
Research Steps
NAMD
GROMACS
CHARMM
Biophysicists prepared molecules on their computer, ran simulations on remote supercomputers, then manually pulled results back for visualization - all this while managing different tools for each step. This fragmented workflow broke focus and slowed their progress.
Primary Research
To better understand workflow friction, I conducted semi-structured interviews with 2 biophysicists from University of Illinois-Urbana Champaign (UIUC).
With the data points received from the interviews, I carried out affinity mapping to identify themes, pain points and define design goals.
Workflow Visibility & Status Tracking
Project & Simulation Management
Tool Fragmentation & Workflow Breaks
Natural Language Interface
FlickAI provides a conversational interface that understands user's everyday language and turns it into simple, guided tasks.
Synthesizing the data points from primary research, 3 key learnings were identified -
Unclear Experiment Status
Scientists can’t easily tell if experiments are running, stuck, or failed without digging into multiple tools.
Poor Project Management
As experiments scale across projects and people, it becomes hard to track ownership, context, and progress.
Scattered Tools slow down work
Manually moving between tools and files shifts focus away from analysis and slows discovery.
Design Goals
Primary research and workflow analysis shaped Cybershuttle’s design goals around clarity, structure, and scalable workflows.
Bring Structure to Complex Research Workflows
Help researchers manage multiple experiments, projects, and collaborators without losing context or ownership as work scales. Organization should reduce complexity, not add to it.
Reduce Fragmentation across Tools and Systems
Minimize context switching by unifying the research workflow and tools into a consolidated platform. Less tool management. More scientific thinking.
Make Experiment State Clear at all Times
Design for instant understanding of whether an experiment is running, completed, or failed, without forcing users to dig through logs or multiple tools.
Design for Scalability without Increasing Complexity
Ensure the system continues to feel manageable as experiments, data, and collaborators grow, without overwhelming the user.
THE SOLUTION
Cybershuttle. A unified workspace for end-to-end molecular simulation work.
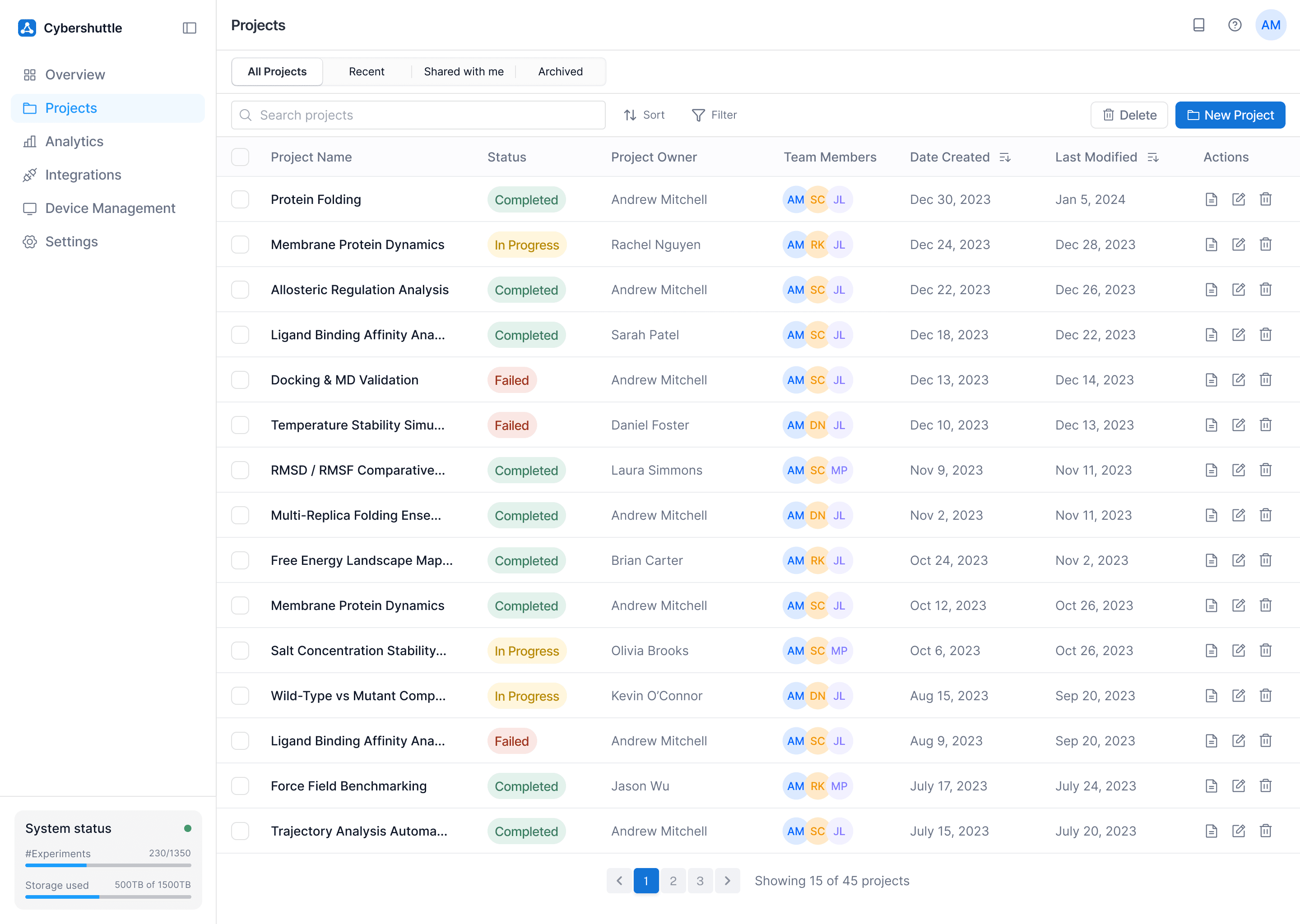
Problem Tackled
Biophysicists struggled to understand the state of their work at a glance.
How
Projects are organized into a single, scannable table with clear status, ownership, and actions.
Why
At-a-glance awareness helps scientists quickly understand progress without reopening tools.
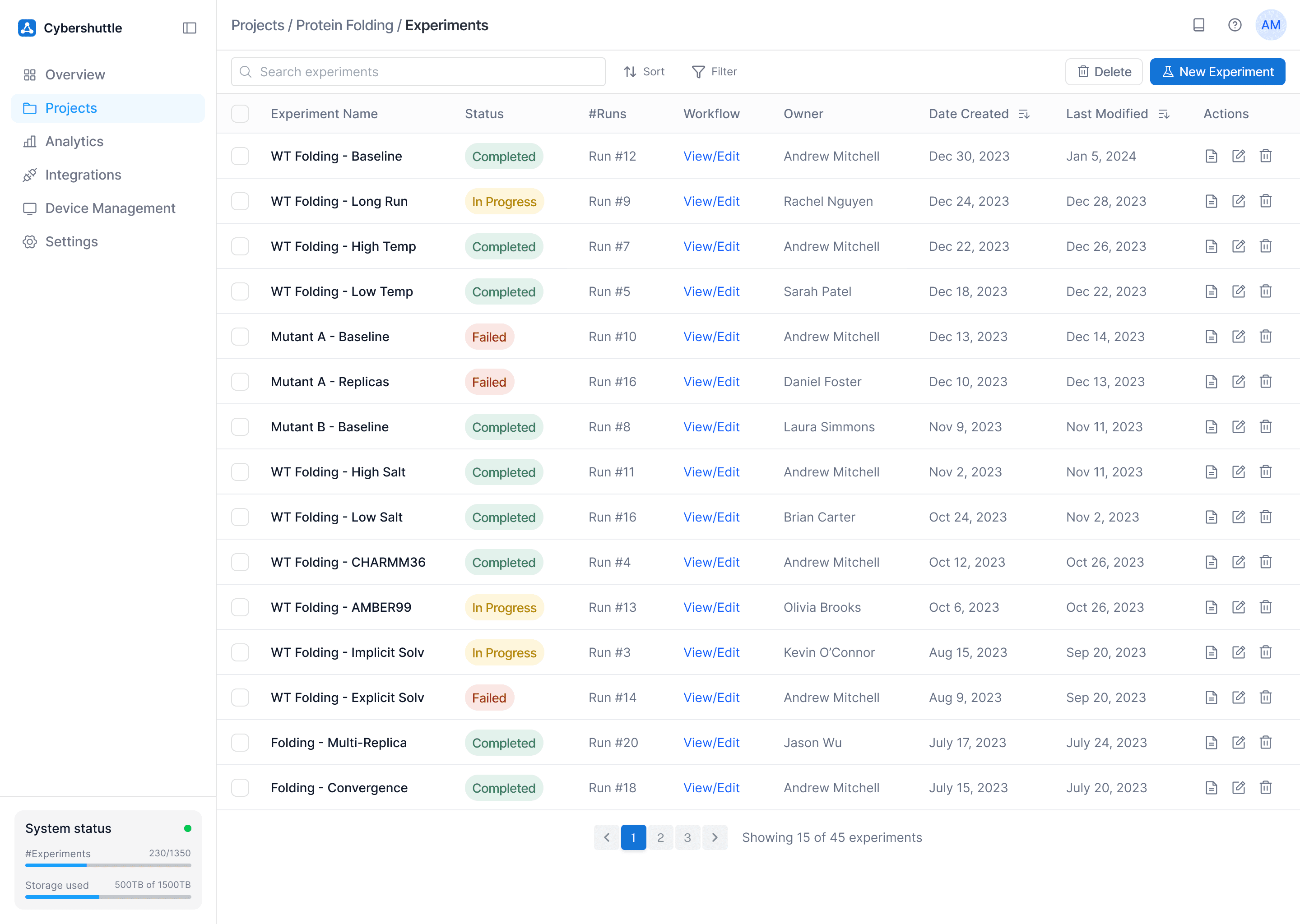
Problem Tackled
Within a project, biophysicists had no easy way to monitor experiments.
How
Experiments are grouped under a project and labeled with status, run count, workflow, and owner.
Why
Since each project has multiple experiments, a heirarchical flow offers a smooth user experience.
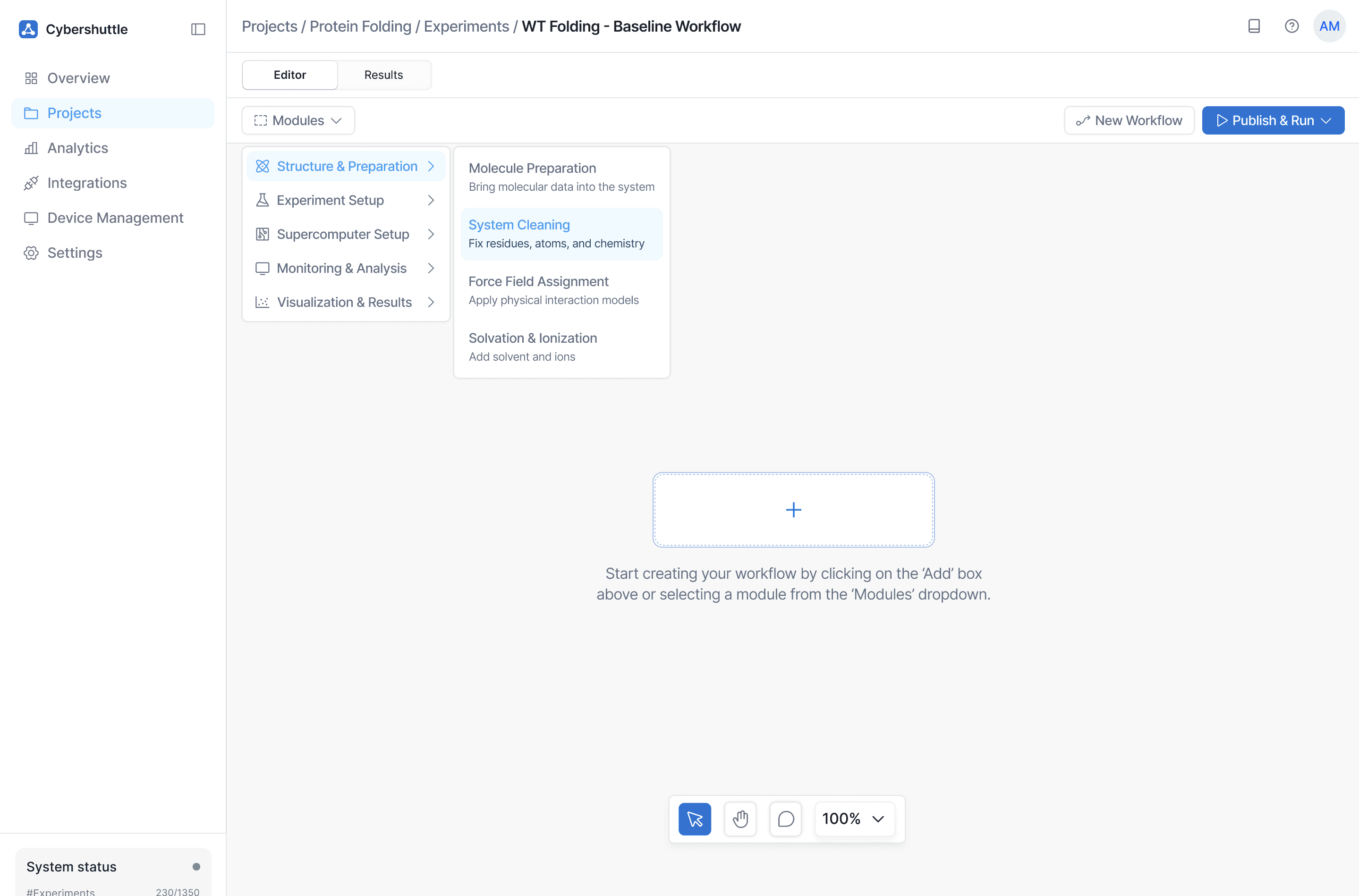
Problem Tackled
Tools dictated the workflow instead of the experiment itself, forcing users to think in terms of software rather than scientific intent.
How
Modules are organized by research actions (e.g., preparation, analysis), not tools.
Why
Designing around intent rather than tooling makes workflows easier to learn, reuse, and explain.
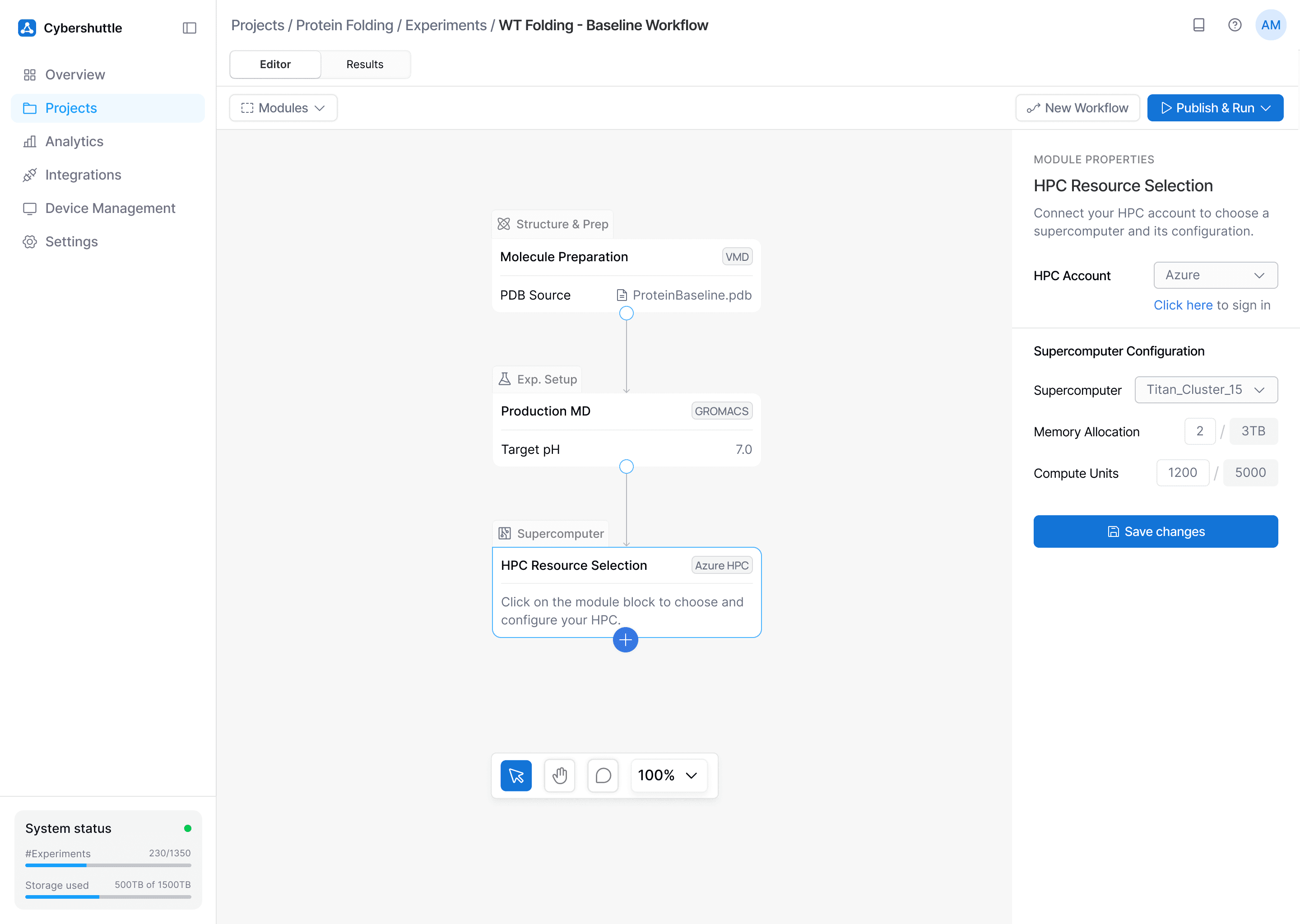
Problem Tackled
Research workflows break down with disconnected tools and configurations across devices.
How
The workflow builder brings tools, steps, and configurations under one unified space, fully customizable, end to end.
Why
A single, coherent system reduces fragmentation while giving biophysicists a bird's eye view of their research processes.
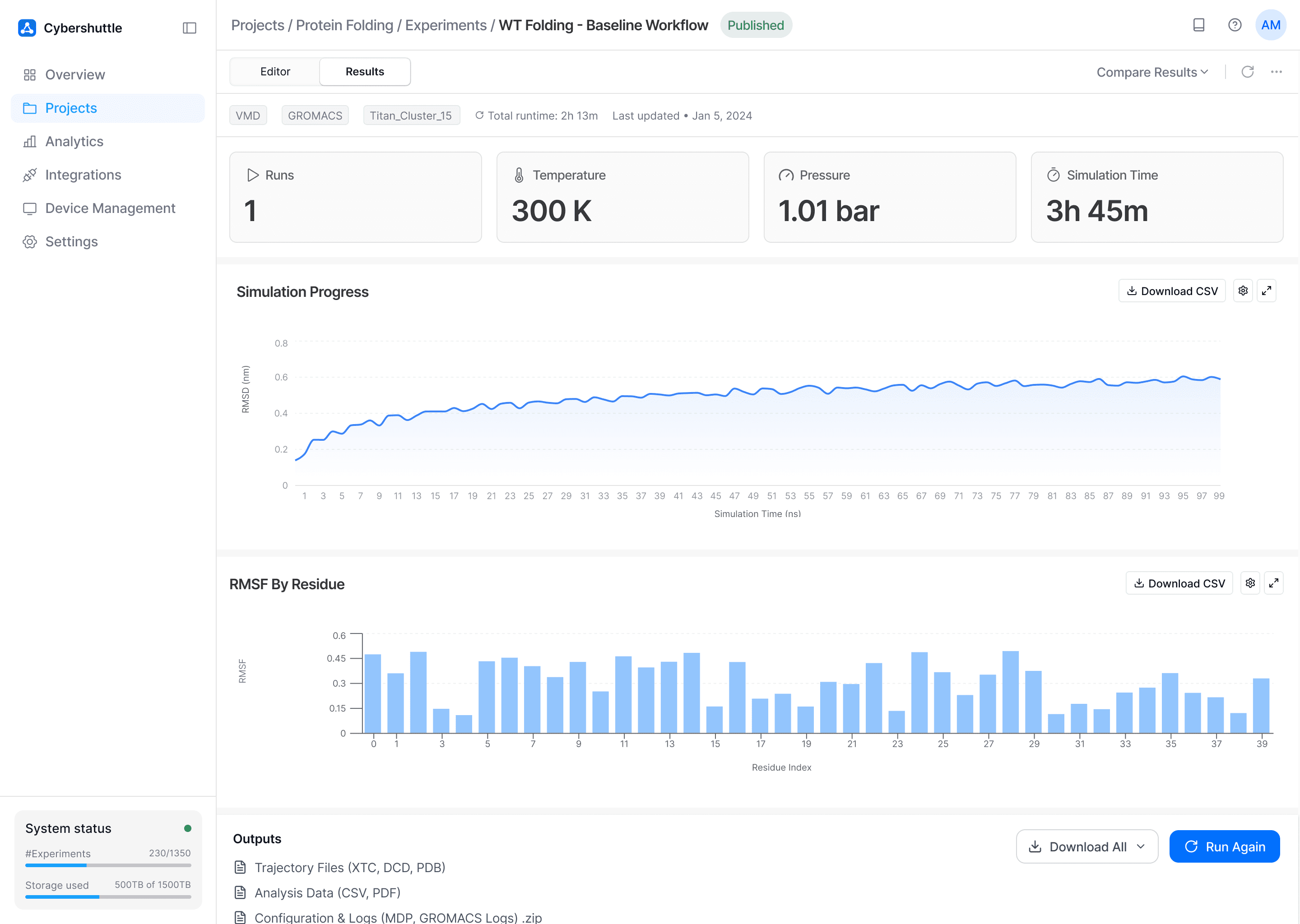
Problem Tackled
Visualization of results required external tools, slowing down analysis and decision making.
How
Simulation metrics, plots, and outputs are surfaced directly alongside the workflow builder.
Why
Keeping results close to the workflow supports faster decision-making and modifications if required.
LEARNINGS
Cybershuttle taught me that unifying tools isn’t enough, you must unify how people think and talk about work.
Speaking the user's language
Biophysicists didn’t think in terms of “using VMD," they thought in terms of their research intent and outcomes. Shifting the interface to be focused on what they want to do - made ownership, comparison, and tracking feel natural.
A unified interface creates shared language, not just shared access
By standardizing how experiments, workflows, and results were represented, Cybershuttle helped teams talk about their work more clearly, even outside the product.
Thoughtful design reduces the need for optional expertise
Before Cybershuttle, deep tool knowledge was often used as a proxy for expertise. A unified system allowed scientists to focus on scientific reasoning, not operational mastery.

